The overall goal of the Data Processing and Analysis Core (DPAC) is to promote a systems biological approach to the study of brain-gut-microbiome (BGM) interactions, with a focus on sex-related differences in these interactions. The Core will apply advanced computational, biostatistical, and bioinformatics approaches to assess the interaction between various levels of biological data (e.g. microbiota, metabolites, sex hormones, multimodal brain and brainstem imaging data, clinical variables (symptom severity, widespread pain) and sex.
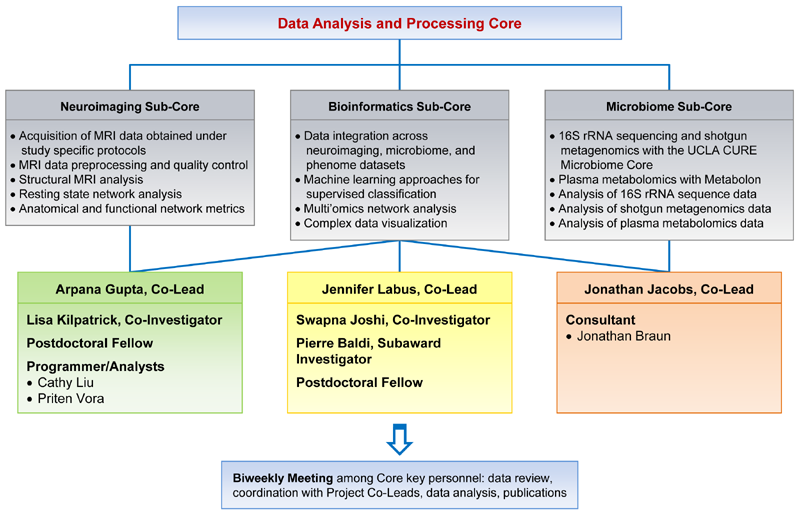
Main components of DPAC
- The gut microbiome (16s rRNA sequencing, shotgun metagenomics, metabolomics)
- Human brain imaging (brain data from advanced multimodal imaging technologies including brainstem imaging, diffusion spectrum imaging, functional magnetic resonance imaging (fMRI), high resolution structural MRI to phenotype the whole brain)
- Multi’omics integration of data using big data and biological systems-based approaches
Specific Aims
- Design and implement protocols for microbiome assessment, and neuroimaging acquisition
- Monitor and ensure quality control of all data collection for all study measures
- Perform advanced microbiome and metabolomic analyses
- Provide quality control, preprocessing and analysis of brain imaging
- Apply multi-omics integration analysis to brain, microbiome, metabolomics, hormone, behavioral and clinical data from individual Projects
- Participate in the interpretation and write up of results along with Project Leads
People
Neuroimaging Sub-Core: Arpana Gupta, PhD (Co-Lead)
Bioinformatics Sub-Core: Jennifer Labus, PhD (Co-Lead)
Microbiome Sub-Core: Jonathan Jacobs, MD, PhD (Co-Lead)

Arpana Gupta, PhD
Co-Director, Neuroimaging and Bioinformatics Core, G. Oppenheimer Center for Neurobiology of Stress and Resilience; Assistant Professor, Vatche and Tamar Manoukian Division of Digestive Diseases, David Geffen School of Medicine at UCLA
Arpana Gupta is an Assistant Professor and Director of the Neuroimaging Core at the UCLA G. Oppenheimer Center of Neurobiology of Stress and Resilience (CNSR); where she specializes in research that investigates the interactions between environmental and biological factors in shaping neurobiological phenotypes associated with stress-based diseases such as obesity and metabolic syndrome. Her current program of research, broadly defined, is based on developing a model that aims to understand the bidirectional interaction of the brain with those in the periphery (immune cells, gut microbiota-related metabolites), and the modification of these interactions by vulnerability or protective factors (adverse life events, sex, race, socioeconomic status [SES], resilience, diet) related to obesity and ingestive behaviors. More recently she has been investigating diet interventions in altering the brain-gut microbiome axis on health and disease. Another main area of interest is sex differences in central responses related to the brain-gut microbiome axis, as well as its relationship to various disease states. She applies advanced multivariate analytic techniques in order to integrate data from multiple neuroimaging sources, inflammatory markers, microbiome and metabolite profiles, and behavioral data, in order to determine the unique variance associated with altered brain gut microbiome axis in specific disorders. In 2016, she received a mentored K23 grant and in 2020 a R03 grant from the NIH NIDDK to investigate the brain-gut microbiome influences in obesity. She has also received funding from the AGA Rome Foundation, Biocodex, and pilot funds from the UCLA CURE/CTSI program.
Publications
http://www.ncbi.nlm.nih.gov/sites/myncbi/1v_1kl872tPQf/bibliography/46222127/public

Jonathan Jacobs, MD, PhD
Assistant Professor-in-Residence, Vatche and Tamar Manoukian Division of Digestive Diseases, David Geffen School of Medicine at UCLA
Dr. Jacobs’ research program focuses on characterizing host-microbiome interactions in patients with gastrointestinal, metabolic, and inflammatory disorders using a combination of human association studies and animal models, including humanized gnotobiotic mice. This evolved initially from several translational microbiome studies he performed investigating the mucosal microbiome and metabolome of inflammatory bowel disease patients compared to healthy family members or unrelated controls, stratified by genetic traits. These studies required the development and refinement of efficient pipelines for 16S ribosomal RNA sequencing, metabolomics, and bioinformatics analysis of multi’omics datasets. He established the Microbiome Core for the UCLA Microbiome Center to offer a range of microbiome-related services to the local research community. This core became affiliated with the UCLA Specialized Center of Research (SCOR) in Neurovisceral Sciences and Women’s Health led by Emeran Mayer and Lin Chang. Through this collaboration, his laboratory assumed responsibility for sample processing, 16S ribosomal RNA sequencing, and bioinformatics analysis for initial SCOR studies on sex differences in the brain-gut-microbiome axis of irritable bowel syndrome (IBS) patients and healthy controls, and their relationship to symptom severity and psychological parameters. This research led to a publication in Microbiome linking microbiome features to brain structural parameters and unpublished work was presented at last year’s Digestive Diseases Week showing that the gut microbiome predicts response of IBS patients to cognitive behavioral therapy.
The current proposal would build upon these collaborations to establish a unique research program within the UCLA Center for Neurovisceral Sciences and Women’s Health dissecting sex differences in brain-gut-microbiome pathways underlying IBS. He serves as co-lead of the Data Processing and Analysis Core, with primary responsibility for microbiome analyses across all three projects and joint responsibility with his co-Leads, Jennifer Labus and Arpana Gupta, for integrative bioinformatics analyses bridging the microbiome, metabolome, neuroimaging, and clinical parameters. This exciting research would draw upon his extensive background in microbiome bioinformatics and experience as Director of the UCLA Microbiome Core.
Publications
http://www.ncbi.nlm.nih.gov/myncbi/browse/collection/48438874

Jennifer Labus, PhD
Director, Biostatistics and Bioinformatics Core, G. Oppenheimer Center for Neurobiology of Stress and Resilience; Adjunct Professor, Vatche and Tamar Manoukian Division of Digestive Diseases, David Geffen School of Medicine at UCLA
Dr. Labus is an Adjunct Professor in the Vatche and Tamar Manoukian Division of Digestive Diseases in the Department of Medicine at University of California, Los Angeles (UCLA). She is the Director of the Integrative Biostatistics and Bioinformatics Core in the G. Oppenheimer Center for Neurobiology of Stress at UCLA and the UCLA Microbiome Center.
Dr Labus is an applied statistician with expertise in biostatistics, bioinformatics, treatment-outcome research, pain neuroscience, multimodal brain imaging, microbiome, metabolomics, and multi-omics integrative analysis. Her current research focused is on determining biological markers of disease, including chronic pain, obesity and Alzheimer’s disease. Using state-or-the-art computational, biostatistical, and bioinformatics approaches, she assesses the interaction between various levels of biological data (e.g., microbiome, metabolomics, immune markers, multimodal brain imaging data) with clinical data. The overall goal of her systems-based approach is to identify and target the key regulators of multi-omics-biological disease-interaction networks in order to understand the underlying pathophysiological mechanisms and provide new targets for treatment.
Dr Labus has made seminal contributions to mapping neural networks underlying visceral pain and elucidating brain-gut –microbiome axis in humans. As a result, she was the recipient of the 2011 Master’s Award for Outstanding Achievement in Basic or Clinical Digestive Sciences, American Gastroenterology Association. Dr Labus has been the recipient of a K08 Career Development award, Effective connectivity of central response in irritable bowel disorder, from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). She has served as the primary investigator on two grants funded by the National Institute of Childhood Health and Human Development (NICHD): R01HD076756 Profiling vulvodynia subtypes based on neurobiological and behavioral endophenotypes and R21HD086737 Deriving novel biomarkers of localized provoked vulvodynia through metabolomics: A biological system-based approach. Labus is a co-investigator on several NIH funded grants, international research collaborations, and is actively involved in mentoring graduate students and postdoctoral fellows.
Publications
http://www.ncbi.nlm.nih.gov/sites/myncbi/1TAcC6itlmG/bibliography/44260598/public/
